Visualizing subcellular structures deep inside the brains of living animals could improve our understanding of how neurons function in their native environment. Thanks to an improved super-resolution microscopy technique developed at Yale University, that dream is now one step closer to reality.
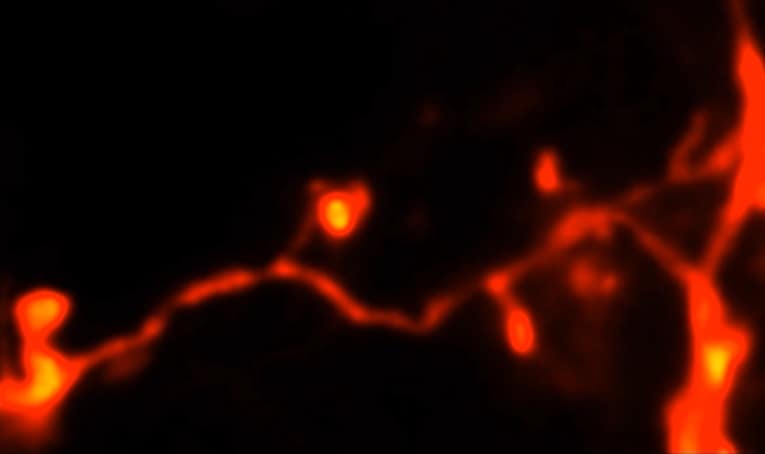
Writing in Optica, the researchers combined stimulated emission depletion (STED) microscopy with two-photon excitation (2PE) to visualize, in three dimensions, the dendritic spines of live mice. These tiny, branch-like protrusions on nerve cells play a major role in neuron-to-neuron communication. Using their 3D-2PE-STED microscope, the researchers observed morphological changes in spines almost 100 μm deep in the brain.
The microscope marks the next generation of 3D-STED technology. Importantly, the technique could offer an in vivo view of several nanoscale structures buried deep within biological tissues.
“The ability to study cellular behaviour in this way is critical to gaining a comprehensive understanding of biological phenomena for biomedical research as well as for pharmaceutical development,” says lead investigator Joerg Bewersdorf.
Capturing the finer details
Designing a microscope that is compatible with in vivo imaging is far more challenging than imaging cells cultured on a glass coverslip. Not only does the light have to navigate through thick, optically dense tissue, but any movement (such as breathing and heartbeat) can create motion artefacts within the image. The net result is poor resolution, particularly in the axial (z) direction.
The 3D-2PE-STED microscope developed by the Bewersdorf Laboratory addresses two key barriers to deep-tissue imaging: light scattering; and optical aberration (where refractive index variations both within the tissue and between the tissue and the objective lens immersion medium create blurry, out-of-focus images). Incorporating 2PE – which uses near-infrared light to generate fluorescent signals in the region-of-interest – reduces light scattering. Meanwhile, adaptive optics and an optimized immersion lens cancel out optical aberrations from the tissue, allowing the light to focus properly.
Before testing their setup in vivo, the researchers benchmarked the super-resolution capabilities of their system against a standard 2PE microscope.
First, they took images of cells cultured on a coverslip. The 3D-2PE-STED system achieved lateral (x–y) and axial resolutions of 70 and 151 nm, respectively – 4.2 and 6.5 times higher than the 2PE microscope. When the researchers tested thicker tissue samples, the resolving power of the 3D-2PE-STED system uncovered structural details within DNA that were lost in the 2PE image.
The full potential of their microscope was then realized 76 μm deep inside the brain of a living mouse. The team successfully tracked the 3D structure of individual spines over three days.
“3D-2PE-STED now provides the means to observe changes [to dendritic spines], and to do so not only in the superficial layers of the brain, but also deeper inside, where more of the interesting connections happen,” explains first author Mary Grace Velasco.
Importantly, the researchers did not see any abnormal changes to the neurons or the mouse’s behaviour as a result of the imaging conditions. They believe that their microscopy technique could help uncover a vast number of structural and cellular relationships found deep within different tissues.